I’ll check it out.
I think caymans list has some extras on it like the weird delta 10 and 6 isomers ive posted the
link in my separations thread.
also they are updating that list regularly.
NIST 2011 x86 torrent found on rutracker
magnet:?xt=urn:btih:66C672056351EACA2607F6A45E10E647FFDEB2E7&tr=http%3A%2F%2Fbt2.t-ru.org%2Fann%3Fmagnet&dn=NIST%2FEPA%2FNIH%20Mass%20Spectral%20Library%202011%20x86%20%5B2011%2C%20ENG%5D
updates for ms search ms interpret and AMDIS Software
https://chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:nist17
It’s only a matter of time. The equipment companies are game. Extractors are game. Chemists are game. Just gotta light this powder keg of competition up with some lit prizes / ways for people to bet. In Vegas imo
Funding it strategically. It’s been on my mind for quite some time. There is definitely enough competitive grrr to make it happen. I don’t care if someone else makes it happen either, I would just like to see it come to fruition.
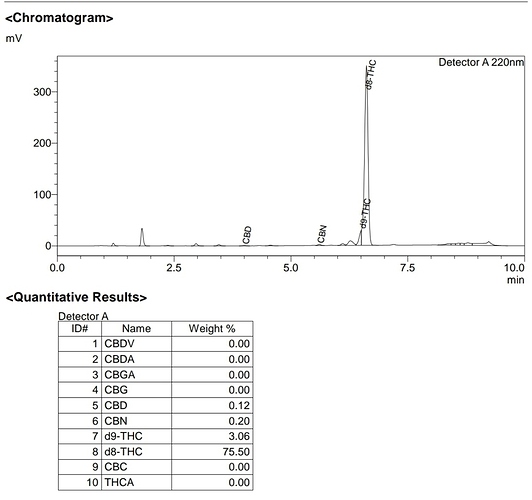
Thank you @Dr_Jebril! And thank you for the HPLC run @MagisterChemist!
I had a conference call with my client and several folks from various @kcalabs locations around the country today on this very subject. We are slowly but surely making progress, thankfully. However, during that call, Steve told me that your CBL elutes between your Δ9 & Δ8 peaks, @kcalabs, yet you said here that, like most methods, your THCs are right next to one another (peaks #12 & #13?)… so is the method you displayed here different than the one he was talking about? In your bluish teal screenshot, peak #12 does not fully return to baseline before #13 begins, leading me to believe this is perhaps a previous method.
As it stands, a lot of the resolution problems between the D8 & D9 peaks seem to be caused by the concentrations of the samples in testing, in combination with the extremely narrow range of retention times at which they both elute.
Because they are so close, in-house testing, for example, can be skewed by even the slightest differences between between manual injection and acquisition start times.
Because they are often too concentrated, even standard dilutions for autosampler injections can cause convolution of the tail and leading slopes of the peaks.
I believe the main reason that @MagisterChemist and @kcalabs (at least stated on the call) are able to achieve “baseline resolution” (full return to baseline) between so many cannabinoids, and especially D9 to D8, even with fairly rapid whole-sample elution times (around 8 and 11 minutes, respectively), is because they are properly diluting them at least 200X for injection!
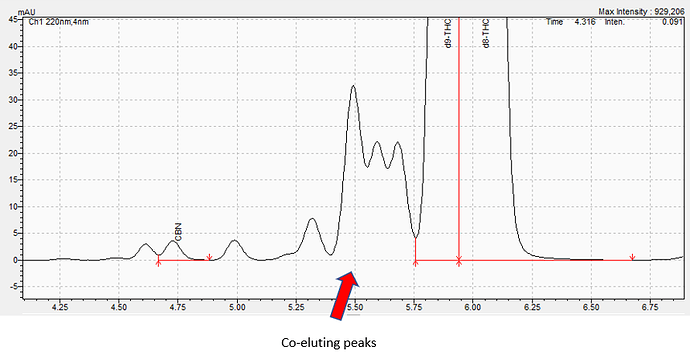
That being said, we may still have a significant issue with certain quinoids co-eluting with the cannabinoids… namely, the finding by @swift.solutions (see Instagram) that HU-331 elutes in the same roi (region/retention of interest/integration) as Δ9-THC! These quinoids are very common byproducts during isomerization reactions, even from pure CBD isolate, especially when lacking sufficient control/exclusion of oxygen.
I am also in talks with Phenomenex regarding screening and implementationaof chiral column chemistries for use in Cannabis analyses. Separations of the enantiomers and diastereomers will be required by CFR under the FDA when it finally happens, and I, for one, would rather have the entire industry prepared for this eventuality, so we don’t have to shut everything down while the labs screen columns and validate methods!
The image below shows the common problem of the day, as I believe @Remedy had also portrayed… and this could be caused by operator error at injection/acquisition start and/or by an overly concentrated sample. Please note I am NOT suggesting “diluting to make things invisible”! Rather, dilution is necessary when specificity/efficiency of the method’s separation of two important analytes is extremely narrow, as it is with the Δ9 & Δ8 THC isomers in these methods.
This shows the shoulder of a D8 peak being “sliced out” to count as D9… which is just the wrong way to do this, certainly without any additional identification of that slice! This sample was likely injected just before acquisition was started on the computer software, making that shoulder edge into the roi of D9:
And the following is an example of a similar chromatogram, slight shoulder on the D8 and all, yet considered to have no D9… which is, unfortunately, ALSO the wrong way to assume the analysis, because, as you might note, this sample was probably just injected right after acquisition was started, pushing that shoulder just downfield enough to keep it out of the D9 roi:
EDIT: Also note the 150 mV differences between the two samples on the y-axis. The 2nd chromatogram is more dilute than the 1st. As you can see, this does not change what analytes can and cannot be seen, as long as it is within reason. The 2nd analysis, had injection/acquisition been started the same way as the 1st, might have been a better analysis than the 1st, having been properly diluted!
Fyi, it is very likely that both of these shoulders are actually HU-331 convoluted with the leading edge of the D8 peak.
@Photon_noir Appreciate you taking this into your own hands! Hopefully more labs can talk to you.
Not sure who you spoke with, but we were not on a call with you yesterday @Photon_noir. Are you confusing us with Kaycha? They have multiple labs across the country, which would make sense. We only have the one location at the moment. Wouldn’t be the first time someone confused “KCA” and “Kaycha.”
Besides the lab intercomparison, I believe we can also further improve in the identitification of the unknowns in the various methods (which turn out to be similar, just in varying proportion), simply based on the existing litrerature, and on the various experiments reported here.
I think HU-331 is in this list, but that it is rather a minor. But I need to improve my understanding. I thought this was the red/purple compound that colors some of such synthesis, or similarly colors pure cbd tinctures exposed to UV. In my gc method, I believe this elutes first, before d9-thcv. The thcs come much later.
There is one compound systematically present in such synthesis, (between 2 and 15%). I strongly suspect that this is either d10-thc, or 8-OH-iso-hhc. I believe this is a competitor to d8. There is another one that is also often there, especially in intermediate steps, or d9 attemps, and it seems intermediate between cbd and d9. This one I suspect to be 9α-OH-hhc.
If oxygen is present, there may be slight formation of CBN and other minor oxidation products (limited to few % altogether).
Some route seem also to diverge to the varine analogues of the above listed compounds.
@Dr_Jebril, you are correct. We’re seeing the slight formation of CBN, too.
@Photon_noir, in highly processed concentrates, we are finding the HPLC-UV/PDA data increasingly more difficult to analyze and, ultimately, trust. There is a lot of co-elution and these “unknowns” make identifying and quantitating known cannabinoids very difficult.
Here is an example:
As you can see, this data is very complex - even for a well-trained analyst. At KCA, we do not rely on one method or one instrument to verify results. We use a variety of instruments, sample preps, dilutions, etc. to ensure that your results are accurate. It is not uncommon for us to inject your sample 4-5 times on two different instruments. At most testing labs, they might inject your sample once on HPLC.
Why do we do this? We want you to trust your results. And we think the only way you can trust your results is to make sure we trust the results ourselves.
We use the mass spec for a variety of reasons but one of the main reasons is mass filtering. The mass spec is great at filtering out interfering masses (which often co-elute on the HPLC-UV/PDA) and improving signal to noise. At KCA, we have a designed a LC-MS/MS method that is a best of both worlds approach – it relies on the mass spec to filter out interfering masses (co-eluting peaks) and designed to separate Δ9 THC and Δ8 THC at the baseline (R > 1.5, based on AOAC acceptance criteria).
@bigbone, I know you wanted to see R > 2, so I’m happy to let you know we achieved a resolution greater than 2 for this method. Here are those chromatograms:
@Photon_noir, if you can clarify who you spoke to yesterday that would be greatly appreciated. Our methods are quite different than what you’re speaking to.
We’ve checked for HU-331 and it has a different mass than d9-THC. We’ve seen HU-331’s mass on the LC-MS/MS, but we do not have a reference standard to confirm.
@kcalabs could you take some cbd isolate, suspend it in pure ethanol, and expose it to ambiant light for few days ? It should turn pink, and yield what I suspect as being hu-331. Then you could detect with the MS, and see if this makes sense.
@Dr_Jebril Would it be UV A, B or C responsible for the reaction?
I don’t know if it is a matter of energy, or also of wavelenght. I would bet UV B are ok. It happens with ambiant sunlight or certain neons tubes, even not directly exposed.
@Photon_noir remember when smokemeout and you were talking shit behind my back about no labeling that shoulder as d9 and saying I was spreading mis information looks like your starting to catch up
A conversation sent to me about photon and smokemeout talking about me…
@Photon_noir you still feel this same way?
@Dr_Jebril we’ll do that. We’ve created our GC-MS/MS method and achieved separation of the isomers on that platform. I’ll see if the scientists have any questions and let them email you directly.
Have you seen the same problems with GC and convolution in the d9 region?
We haven’t run samples on the GC just yet, so we have yet to see whether or not the same problems occur. We have separated d9, d8, exo, d10, and d6a,10a so far.
Using an hp-5 column, you get easily d9 separated from d8, but you get cbc and cbd coeluting. Using an hp35- column, you fix this.
MY MISTAKE, @kcalabs!!! That is exactly the case of mistaken identity! Your logos look very similar at a glance, too, especially when tiny on a phone screen! I apologize, and I appreciate your understanding! ![]()
My experience with Kaycha has been bad; not able to identify various cannabinoids properly. They’re on my do not ever use list. I don’t pay for analytics to be constantly wrong and that’s what I’ve found with Kaycha.
@kcalabs on the other hand is at exactly the opposite end of the spectrum. If the COA is from KCALabs, the product tested has immediate credibility to us.